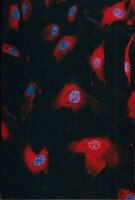
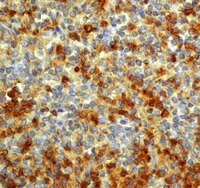
GSI promotes vincristine-induced apoptosis by enhancing multi-polar spindle formation.
Singh, A; Zapata, MC; Choi, YS; Yoon, SO
Cell cycle (Georgetown, Tex.)
13
157-66
2014
Show Abstract
Gamma secretase inhibitors (GSI), cell-permeable small-molecule inhibitors of gamma secretase activity, had been originally developed for the treatment of Alzheimer disease. In recent years, it has been exploited in cancer research to inhibit Notch signaling that is aberrantly activated in various cancers. We previously found that GSI could synergize with anti-microtubule agent, vincristine (VCR) in a Notch-independent manner. Here, we delineate the underlying cell cycle-related mechanism using HeLa cells, which have strong mitotic checkpoints. GSI enhanced VCR-induced cell death, although GSI alone did not affect cell viability at all. GSI augmented VCR-induced mitotic arrest in a dose-dependent manner, which was preceded by apoptotic cell death, as shown by an increase in Annexin V-positive and caspase-positive cell population. Furthermore, GSI amplified multi-polar spindle formation triggered by VCR. Altogether, we show the evidence that GSI enhances VCR-induced apoptosis in HeLa cells via multi-polar mitotic spindle formation, independent of Notch signaling. These data suggest that one or more GS substrates, yet to be identified, in a post-GS processed form, may play a role in maintaining functional centrosomes/mitotic spindles. More significantly, the synergistic effect of GSI in combination with VCR could be exploited in clinical setting to improve the efficacy of VCR. | | 24200971
 |
Activation of the NOTCH pathway in head and neck cancer.
Sun, W; Gaykalova, DA; Ochs, MF; Mambo, E; Arnaoutakis, D; Liu, Y; Loyo, M; Agrawal, N; Howard, J; Li, R; Ahn, S; Fertig, E; Sidransky, D; Houghton, J; Buddavarapu, K; Sanford, T; Choudhary, A; Darden, W; Adai, A; Latham, G; Bishop, J; Sharma, R; Westra, WH; Hennessey, P; Chung, CH; Califano, JA
Cancer research
74
1091-104
2014
Show Abstract
NOTCH1 mutations have been reported to occur in 10% to 15% of head and neck squamous cell carcinomas (HNSCC). To determine the significance of these mutations, we embarked upon a comprehensive study of NOTCH signaling in a cohort of 44 HNSCC tumors and 25 normal mucosal samples through a set of expression, copy number, methylation, and mutation analyses. Copy number increases were identified in NOTCH pathway genes, including the NOTCH ligand JAG1. Gene set analysis defined a differential expression of the NOTCH signaling pathway in HNSCC relative to normal tissues. Analysis of individual pathway-related genes revealed overexpression of ligands JAG1 and JAG2 and receptor NOTCH3. In 32% of the HNSCC examined, activation of the downstream NOTCH effectors HES1/HEY1 was documented. Notably, exomic sequencing identified 5 novel inactivating NOTCH1 mutations in 4 of the 37 tumors analyzed, with none of these tumors exhibiting HES1/HEY1 overexpression. Our results revealed a bimodal pattern of NOTCH pathway alterations in HNSCC, with a smaller subset exhibiting inactivating NOTCH1 receptor mutations but a larger subset exhibiting other NOTCH1 pathway alterations, including increases in expression or gene copy number of the receptor or ligands as well as downstream pathway activation. Our results imply that therapies that target the NOTCH pathway may be more widely suitable for HNSCC treatment than appreciated currently. | | 24351288
 |
Tumor-suppressive activity of Lunatic Fringe in prostate through differential modulation of Notch receptor activation.
Zhang, S; Chung, WC; Wu, G; Egan, SE; Xu, K
Neoplasia (New York, N.Y.)
16
158-67
2014
Show Abstract
Elevated Notch ligand and receptor expression has been associated with aggressive forms of prostate cancer, suggesting a role for Notch signaling in regulation of prostate tumor initiation and progression. Here, we report a critical role for Lunatic Fringe (Lfng), which encodes an O-fucosylpeptide 3-ß-N-acetylglucosaminyltransferase known to modify epidermal growth factor repeats of Notch receptor proteins, in regulation of prostate epithelial differentiation and proliferation, as well as in prostate tumor suppression. Deletion of Lfng in mice caused altered Notch activation in the prostate, associated with elevated accumulation of Notch1, Notch2, and Notch4 intracellular domains, decreased levels of the putative Notch3 intracellular fragment, as well as increased expression of Hes1, Hes5, and Hey2. Loss of Lfng resulted in expansion of the basal layer, increased proliferation of both luminal and basal cells, and ultimately, prostatic intraepithelial neoplasia. The Lfng-null prostate showed down-regulation of prostatic tumor suppressor gene NKX3.1 and increased androgen receptor expression. Interestingly, expression of LFNG and NKX3.1 were positively correlated in publically available human prostate cancer data sets. Knockdown of LFNG in DU-145 prostate cancer cells led to expansion of CD44(+)CD24(-) and CD49f(+)CD24(-) stem/progenitor-like cell population associated with enhanced prostatosphere-forming capacity. Taken together, these data revealed a tumor-suppressive role for Lfng in the prostate through differential regulation of Notch signaling. | Western Blotting | 24709423
 |
Crosstalk between glioma-initiating cells and endothelial cells drives tumor progression.
Jeon, HM; Kim, SH; Jin, X; Park, JB; Kim, SH; Joshi, K; Nakano, I; Kim, H
Cancer research
74
4482-92
2014
Show Abstract
Glioma-initiating cells (GIC), which reside within the perivascular microenvironment to maintain self-renewal capacity, are responsible for glioblastoma initiation, progression, and recurrence. However, the molecular mechanisms controlling crosstalk between GICs and endothelial cells are poorly understood. Here, we report that, in both GICs and endothelial cells, platelet-derived growth factor (PDGF)-driven activation of nitric oxide (NO) synthase increases NO-dependent inhibitor of differentiation 4 (ID4) expression, which in turn promotes JAGGED1-NOTCH activity through suppression of miR129 that specifically represses JAGGED1 suppression. This signaling axis promotes tumor progression along with increased GIC self-renewal and growth of tumor vasculature in the xenograft tumors, which is dramatically suppressed by NOTCH inhibitor. ID4 levels correlate positively with NOS2 (NO synthase-2), HES1, and HEY1 and negatively with miR129 in primary GICs. Thus, targeting the PDGF-NOS-ID4-miR129 axis and NOTCH activity in the perivascular microenvironment might serve as an efficacious therapeutic modality for glioblastoma. | Immunofluorescence | 24962027
 |
Notch-induced transcription factors are predictive of survival and 5-fluorouracil response in colorectal cancer patients.
Candy, PA; Phillips, MR; Redfern, AD; Colley, SM; Davidson, JA; Stuart, LM; Wood, BA; Zeps, N; Leedman, PJ
British journal of cancer
109
1023-30
2013
Show Abstract
The purpose of this study was to evaluate the expression of Notch-induced transcription factors (NTFs) HEY1, HES1 and SOX9 in colorectal cancer (CRC) patients to determine their clinicopathologic and prognostic significance.Levels of HEY1, HES1 and SOX9 protein were measured by immunohistochemistry in a nonmalignant and malignant tissue microarray of 441 CRC patients, and the findings correlated with pathologic, molecular and clinical variables.The NTFs HEY1, HES1 and SOX9 were overexpressed in tumours relative to colonic mucosa (OR=3.44, Pless than 0.0001; OR=7.40, Pless than 0.0001; OR=4.08 Pless than 0.0001, respectively). HEY1 overexpression was a negative prognostic factor for all CRC patients (HR=1.29, P=0.023) and strongly correlated with perineural and vascular invasion and lymph node (LN) metastasis. In 5-fluorouracil (5-FU)-treated patients, the tumour overexpression of SOX9 correlated with markedly poorer survival (HR=8.72, P=0.034), but had no predictive effect in untreated patients (HR=0.70, P=0.29). When HEY1, HES1 and SOX9 expression were combined to predict survival with chemotherapy, in treated patients there was an additive increase in the risk of death with each NTF overexpressed (HR=2.09, P=0.01), but no prognostic import in the untreated patient group (HR=0.74, P=0.19).The present study is the first to discover that HEY1 overexpression correlates with poorer outcome in CRC, and NTF expression is predictive of CRC patient survival with 5-FU chemotherapy. If confirmed in future studies, testing of NTF expression has the potential to enter routine pathological practice for the selection of patients to undergo chemotherapy alone or in combination with Notch inhibitors. | | 23900217
 |
Midkine promotes neuroblastoma through Notch2 signaling.
Kishida, S; Mu, P; Miyakawa, S; Fujiwara, M; Abe, T; Sakamoto, K; Onishi, A; Nakamura, Y; Kadomatsu, K
Cancer research
73
1318-27
2013
Show Abstract
Midkine is a heparin-binding growth factor highly expressed in various cancers, including neuroblastoma, the most common extracranial pediatric solid tumor. Prognosis of patients with neuroblastoma in which MYCN is amplified remains particularly poor. In this study, we used a MYCN transgenic model for neuroblastoma in which midkine is highly expressed in precancerous lesions of sympathetic ganglia. Genetic ablation of midkine in this model delayed tumor formation and reduced tumor incidence. Furthermore, an RNA aptamer that specifically bound midkine suppressed the growth of neuroblastoma cells in vitro and in vivo in tumor xenografts. In precancerous lesions, midkine-deficient MYCN transgenic mice exhibited defects in activation of Notch2, a candidate midkine receptor, and expression of the Notch target gene HES1. Similarly, RNA aptamer-treated tumor xenografts also showed attenuation of Notch2-HES1 signaling. Our findings establish a critical role for the midkine-Notch2 signaling axis in neuroblastoma tumorigenesis, which implicates new strategies to treat neuroblastoma. | Immunohistochemistry | 23243020
 |
Mutant p53-Notch1 Signaling Axis Is Involved in Curcumin-Induced Apoptosis of Breast Cancer Cells.
Bae, YH; Ryu, JH; Park, HJ; Kim, KR; Wee, HJ; Lee, OH; Jang, HO; Bae, MK; Kim, KW; Bae, SK
The Korean journal of physiology & pharmacology : official journal of the Korean Physiological Society and the Korean Society of Pharmacology
17
291-7
2013
Show Abstract
Notch1 has been reported to be highly expressed in triple-negative and other subtypes of breast cancer. Mutant p53 (R280K) is overexpressed in MDA-MB-231 triple-negative human breast cancer cells. The present study aimed to determine whether the mutant p53 can be a potent transcriptional activator of the Notch1 in MDA-MB-231 cells, and explore the role of this mutant p53-Notch1 axis in curcumin-induced apoptosis. We found that curcumin treatment resulted in an induction of apoptosis in MDA-MB-231 cells, together with downregulation of Notch1 and its downstream target, Hes1. This reduction in Notch1 expression was determined to be due to the decreased activity of endogenous mutant p53. We confirmed the suppressive effect of curcumin on Notch1 transcription by performing a Notch1 promoter-driven reporter assay and identified a putative p53-binding site in the Notch1 promoter by EMSA and chromatin immunoprecipitation analysis. Overexpression of mutant p53 increased Notch1 promoter activity, whereas knockdown of mutant p53 by small interfering RNA suppressed Notch1 expression, leading to the induction of cellular apoptosis. Moreover, curcumin-induced apoptosis was further enhanced by the knockdown of Notch1 or mutant p53, but it was decreased by the overexpression of active Notch1. Taken together, our results demonstrate, for the first time, that Notch1 is a transcriptional target of mutant p53 in breast cancer cells and suggest that the targeting of mutant p53 and/or Notch1 may be combined with a chemotherapeutic strategy to improve the response of breast cancer cells to curcumin. | | 23946688
 |
A mouse model for inducible overexpression of Prdm14 results in rapid-onset and highly penetrant T-cell acute lymphoblastic leukemia (T-ALL).
Carofino, BL; Ayanga, B; Justice, MJ
Disease models & mechanisms
6
1494-506
2013
Show Abstract
PRDM14 functions in embryonic stem cell (ESC) maintenance to promote the expression of pluripotency-associated genes while suppressing differentiation genes. Expression of PRDM14 is tightly regulated and typically limited to ESCs and primordial germ cells; however, aberrant expression is associated with tumor initiation in a wide variety of human cancers, including breast cancer and leukemia. Here, we describe the generation of a Cre-recombinase-inducible mouse model for the spatial and temporal control of Prdm14 misexpression [ROSA26 floxed-stop Prdm14 (R26PR)]. When R26PR is mated to either of two Cre lines, Mx1-cre or MMTV-cre, mice develop early-onset T-cell acute lymphoblastic leukemia (T-ALL) with median overall survival of 41 and 64 days for R26PR;Mx1-cre and R26PR;MMTV-cre, respectively. T-ALL is characterized by the accumulation of immature single-positive CD8 cells and their widespread infiltration. Leukemia is preceded by a dramatic expansion of cells resembling hematopoietic stem cells and lymphoid-committed progenitors prior to disease onset, accompanied by a blockage in B-cell differentiation at the early pro-B stage. Rapid-onset PRDM14-induced T-ALL requires factors that are present in stem and progenitor cells: R26PR;dLck-cre animals, which express Prdm14 starting at the double-positive stage of thymocyte development, do not develop disease. PRDM14-induced leukemic cells contain high levels of activated NOTCH1 and downstream NOTCH1 targets, including MYC and HES1, and are sensitive to pharmacological inhibition of NOTCH1 with the γ-secretase inhibitor DAPT. Greater than 50% of human T-ALLs harbor activating mutations in NOTCH1; thus, our model carries clinically relevant molecular aberrations. The penetrance, short latency and involvement of the NOTCH1 pathway will make this hematopoietic R26PR mouse model ideal for future studies on disease initiation, relapse and novel therapeutic drug combinations. Furthermore, breeding R26PR to additional Cre lines will allow for the continued development of novel cancer models. | | 24046360
 |
An aquaporin 3-notch1 axis in keratinocyte differentiation and inflammation.
Guo, L; Chen, H; Li, Y; Zhou, Q; Sui, Y
PloS one
8
e80179
2013
Show Abstract
Aquaporin 3 (AQP3) is an aquaglyceroporin which transports water, glycerol and small solutes across the plasma membrane. Its functions are not limited to fluid transport but also involve the regulation of cell proliferation, migration, skin hydration, wound healing and tumorigenesis. While AQP3 has been reported to play an important role in keratinocyte proliferation, its role in differentiation remains controversial. Our study demonstrated that the expression of AQP3 was regulated during differentiation and that it participated in keratinocyte differentiation control. We further revealed that AQP3 was a transcriptional target of Notch signaling, a critical pathway regulating keratinocyte differentiation and tumor suppression, and it regulated differentiation through a reciprocal negative feedback loop with Notch1. When the expression level of AQP3 was elevated, impaired barrier integrity and increased pro-inflammatory cytokine production ensued, mimicking the pathological conditions in Notch deficient mice and in atopic dermatitis. Dysregulation of AQP3 and Notch receptors has been reported in several skin diseases, including skin cancer. Our discovery of the novel AQP3-Notch1 axis may provide insight into epidermal homeostasis control and possible translational applications, including its potential use as a biomarker for molecular diagnosis in environmental studies. | Western Blotting | 24260356
 |
Notch-mediated induction of N-cadherin and α9-integrin confers higher invasive phenotype on rhabdomyosarcoma cells.
Masià, A; Almazán-Moga, A; Velasco, P; Reventós, J; Torán, N; Sánchez de Toledo, J; Roma, J; Gallego, S
British journal of cancer
107
1374-83
2012
Show Abstract
Rhabdomyosarcoma (RMS) is the commonest type of soft-tissue sarcoma in children. Patients with metastatic RMS continue to have very poor prognosis. Recently, several works have demonstrated a connection between Notch pathway activation and the regulation of cell motility and invasiveness. However, the molecular mechanisms of this possible relationship remain unclear.The Notch pathway was manipulated pharmacologically and genetically. The mRNA changes were analysed by quantitative PCR and protein variations by western blot and immunofluorescence. Finally, the capabilities of RMS cells to adhere, heal a wound and invade were assessed in the presence of neuronal cadherin (N-cadherin)- and α9-integrin-blocking antibodies.Cells treated with γ-secretase inhibitor showed lower adhesion capability and downregulation of N-cadherin and α9-integrin. Genetic manipulation of the Notch pathway led to concomitant variations in N-cadherin and α9-integrin. Treatment with anti-N-cadherin-blocking antibody rendered marked inhibition of cell adhesion and motility, while anti-α9-integrin-blocking antibody exerted a remarkable effect on cell adhesion and invasiveness.Neuronal cadherin and α9-integrin are postulated as leading actors in the association between the Notch pathway and promotion of cell adhesion, motility and invasion, pointing to these proteins and the Notch pathway itself as interesting putative targets for new molecular therapies against metastases in RMS. | | 22976797
 |