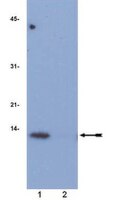
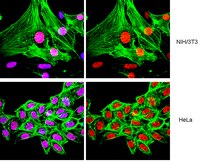
Effects of ethylene glycol monomethyl ether and its metabolite, 2-methoxyacetic acid, on organogenesis stage mouse limbs in vitro.
Dayan, C; Hales, BF
Birth defects research. Part B, Developmental and reproductive toxicology
101
254-61
2014
Show Abstract
Exposure to ethylene glycol monomethyl ether (EGME), a glycol ether compound found in numerous industrial products, or to its active metabolite, 2-methoxyacetic acid (2-MAA), increases the incidence of developmental defects. Using an in vitro limb bud culture system, we tested the hypothesis that the effects of EGME on limb development are mediated by 2-MAA-induced alterations in acetylation programming. Murine gestation day 12 embryonic forelimbs were exposed to 3, 10, or 30 mM EGME or 2-MAA in culture for 6 days to examine effects on limb morphology; limbs were cultured for 1 to 24 hr to monitor effects on the acetylation of histones (H3K9 and H4K12), a nonhistone protein, p53 (p53K379), and markers for cell cycle arrest (p21) and apoptosis (cleaved caspase-3). EGME had little effect on limb morphology and no significant effects on the acetylation of histones or p53 or on biomarkers for cell cycle arrest or apoptosis. In contrast, 2-MAA exposure resulted in a significant concentration-dependent increase in limb abnormalities. 2-MAA induced the hyperacetylation of histones H3K9Ac and H4K12Ac at all concentrations tested (3, 10, and 30 mM). Exposure to 10 or 30 mM 2-MAA significantly increased acetylation of p53 at K379, p21 expression, and caspase-3 cleavage. Thus, 2-MAA, the proximate metabolite of EGME, disrupts limb development in vitro, modifies acetylation programming, and induces biomarkers of cell cycle arrest and apoptosis. | Western Blotting | | 24798094
 |
In vivo imaging of histone deacetylases (HDACs) in the central nervous system and major peripheral organs.
Wang, C; Schroeder, FA; Wey, HY; Borra, R; Wagner, FF; Reis, S; Kim, SW; Holson, EB; Haggarty, SJ; Hooker, JM
Journal of medicinal chemistry
57
7999-8009
2014
Show Abstract
Epigenetic enzymes are now targeted to treat the underlying gene expression dysregulation that contribute to disease pathogenesis. Histone deacetylases (HDACs) have shown broad potential in treatments against cancer and emerging data supports their targeting in the context of cardiovascular disease and central nervous system dysfunction. Development of a molecular agent for non-invasive imaging to elucidate the distribution and functional roles of HDACs in humans will accelerate medical research and drug discovery in this domain. Herein, we describe the synthesis and validation of an HDAC imaging agent, [(11)C]6. Our imaging results demonstrate that this probe has high specificity, good selectivity, and appropriate kinetics and distribution for imaging HDACs in the brain, heart, kidney, pancreas, and spleen. Our findings support the translational potential for [(11)C]6 for human epigenetic imaging. | | | 25203558
 |
A selective HDAC 1/2 inhibitor modulates chromatin and gene expression in brain and alters mouse behavior in two mood-related tests.
Schroeder, FA; Lewis, MC; Fass, DM; Wagner, FF; Zhang, YL; Hennig, KM; Gale, J; Zhao, WN; Reis, S; Barker, DD; Berry-Scott, E; Kim, SW; Clore, EL; Hooker, JM; Holson, EB; Haggarty, SJ; Petryshen, TL
PloS one
8
e71323
2013
Show Abstract
Psychiatric diseases, including schizophrenia, bipolar disorder and major depression, are projected to lead global disease burden within the next decade. Pharmacotherapy, the primary--albeit often ineffective--treatment method, has remained largely unchanged over the past 50 years, highlighting the need for novel target discovery and improved mechanism-based treatments. Here, we examined in wild type mice the impact of chronic, systemic treatment with Compound 60 (Cpd-60), a slow-binding, benzamide-based inhibitor of the class I histone deacetylase (HDAC) family members, HDAC1 and HDAC2, in mood-related behavioral assays responsive to clinically effective drugs. Cpd-60 treatment for one week was associated with attenuated locomotor activity following acute amphetamine challenge. Further, treated mice demonstrated decreased immobility in the forced swim test. These changes are consistent with established effects of clinical mood stabilizers and antidepressants, respectively. Whole-genome expression profiling of specific brain regions (prefrontal cortex, nucleus accumbens, hippocampus) from mice treated with Cpd-60 identified gene expression changes, including a small subset of transcripts that significantly overlapped those previously reported in lithium-treated mice. HDAC inhibition in brain was confirmed by increased histone acetylation both globally and, using chromatin immunoprecipitation, at the promoter regions of upregulated transcripts, a finding consistent with in vivo engagement of HDAC targets. In contrast, treatment with suberoylanilide hydroxamic acid (SAHA), a non-selective fast-binding, hydroxamic acid HDAC 1/2/3/6 inhibitor, was sufficient to increase histone acetylation in brain, but did not alter mood-related behaviors and had dissimilar transcriptional regulatory effects compared to Cpd-60. These results provide evidence that selective inhibition of HDAC1 and HDAC2 in brain may provide an epigenetic-based target for developing improved treatments for mood disorders and other brain disorders with altered chromatin-mediated neuroplasticity. | Western Blotting | Mouse | 23967191
 |
Crebinostat: a novel cognitive enhancer that inhibits histone deacetylase activity and modulates chromatin-mediated neuroplasticity.
Fass, DM; Reis, SA; Ghosh, B; Hennig, KM; Joseph, NF; Zhao, WN; Nieland, TJ; Guan, JS; Kuhnle, CE; Tang, W; Barker, DD; Mazitschek, R; Schreiber, SL; Tsai, LH; Haggarty, SJ
Neuropharmacology
64
81-96
2013
Show Abstract
Long-term memory formation is known to be critically dependent upon de novo gene expression in the brain. As a consequence, pharmacological enhancement of the transcriptional processes mediating long-term memory formation provides a potential therapeutic strategy for cognitive disorders involving aberrant neuroplasticity. Here we focus on the identification and characterization of small molecule inhibitors of histone deacetylases (HDACs) as enhancers of CREB (cAMP response element-binding protein)-regulated transcription and modulators of chromatin-mediated neuroplasticity. Using a CREB reporter gene cell line, we screened a library of small molecules structurally related to known HDAC inhibitors leading to the identification of a probe we termed crebinostat that produced robust activation of CREB-mediated transcription. Further characterization of crebinostat revealed its potent inhibition of the deacetylase activity of recombinant class I HDACs 1, 2, 3, and class IIb HDAC6, with weaker inhibition of the class I HDAC8 and no significant inhibition of the class IIa HDACs 4, 5, 7, and 9. In cultured mouse primary neurons, crebinostat potently induced acetylation of both histone H3 and histone H4 as well as enhanced the expression of the CREB target gene Egr1 (early growth response 1). Using a hippocampus-dependent, contextual fear conditioning paradigm, mice systemically administered crebinostat for a ten day time period exhibited enhanced memory. To gain insight into the molecular mechanisms of memory enhancement by HDAC inhibitors, whole genome transcriptome profiling of cultured mouse primary neurons treated with crebinostat, combined with bioinformatic analyses of CREB-target genes, was performed revealing a highly connected protein-protein interaction network reflecting modules of genes important to synaptic structure and plasticity. Consistent with these findings, crebinostat treatment increased the density of synapsin-1 punctae along dendrites in cultured neurons. Finally, crebinostat treatment of cultured mouse primary neurons was found to upregulate Bdnf (brain-derived neurotrophic factor) and Grn (granulin) and downregulate Mapt (tau) gene expression-genes implicated in aging-related cognitive decline and cognitive disorders. Taken together, these results demonstrate that crebinostat provides a novel probe to modulate chromatin-mediated neuroplasticity and further suggests that pharmacological optimization of selective of HDAC inhibitors may provide an effective therapeutic approach for human cognitive disorders. This article is part of a Special Issue entitled 'Cognitive Enhancers'. | | | 22771460
 |
Fasting and high-fat diet alter histone deacetylase expression in the medial hypothalamus.
Funato, H; Oda, S; Yokofujita, J; Igarashi, H; Kuroda, M
PloS one
6
e18950
2011
Show Abstract
Increasing attention is now being given to the epigenetic regulation of animal and human behaviors including the stress response and drug addiction. Epigenetic factors also influence feeding behavior and metabolic phenotypes, such as obesity and insulin sensitivity. In response to fasting and high-fat diets, the medial hypothalamus changes the expression of neuropeptides regulating feeding, metabolism, and reproductive behaviors. Histone deacetylases (HDACs) are involved in the epigenetic control of gene expression and alter behavior in response to a variety of environmental factors. Here, we examined the expression of HDAC family members in the medial hypothalamus of mice in response to either fasting or a high-fat diet. In response to fasting, HDAC3 and -4 expression levels increased while HDAC10 and -11 levels decreased. Four weeks on a high-fat diet resulted in the increased expression of HDAC5 and -8. Moreover, fasting decreased the number of acetylated histone H3- and acetylated histone H4-positive cells in the ventrolateral subdivision of the ventromedial hypothalamus. Therefore, HDACs may be implicated in altered gene expression profiles in the medial hypothalamus under different metabolic states. Full Text Article | Immunohistochemistry | | 21526203
 |
Mass spectrometry analysis of the variants of histone H3 and H4 of soybean and their post-translational modifications.
Wu, T; Yuan, T; Tsai, SN; Wang, C; Sun, SM; Lam, HM; Ngai, SM
BMC plant biology
9
98
2009
Show Abstract
Histone modifications and histone variants are of importance in many biological processes. To understand the biological functions of the global dynamics of histone modifications and histone variants in higher plants, we elucidated the variants and post-translational modifications of histones in soybean, a legume plant with a much bigger genome than that of Arabidopsis thaliana.In soybean leaves, mono-, di- and tri-methylation at Lysine 4, Lysine 27 and Lysine 36, and acetylation at Lysine 14, 18 and 23 were detected in HISTONE H3. Lysine 27 was prone to being mono-methylated, while tri-methylation was predominant at Lysine 36. We also observed that Lysine 27 methylation and Lysine 36 methylation usually excluded each other in HISTONE H3. Although methylation at HISTONE H3 Lysine 79 was not reported in A. thaliana, mono- and di-methylated HISTONE H3 Lysine 79 were detected in soybean. Besides, acetylation at Lysine 8 and 12 of HISTONE H4 in soybean were identified. Using a combination of mass spectrometry and nano-liquid chromatography, two variants of HISTONE H3 were detected and their modifications were determined. They were different at positions of A31F41S87S90 (HISTONE variant H3.1) and T31Y41H87L90 (HISTONE variant H3.2), respectively. The methylation patterns in these two HISTONE H3 variants also exhibited differences. Lysine 4 and Lysine 36 methylation were only detected in HISTONE H3.2, suggesting that HISTONE variant H3.2 might be associated with actively transcribing genes. In addition, two variants of histone H4 (H4.1 and H4.2) were also detected, which were missing in other organisms. In the histone variant H4.1 and H4.2, the amino acid 60 was isoleucine and valine, respectively.This work revealed several distinct variants of soybean histone and their modifications that were different from A. thaliana, thus providing important biological information toward further understanding of the histone modifications and their functional significance in higher plants. | Western Blotting | | 19643030
 |
RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation.
Carey, Michael, et al.
Mol. Cell, 24: 481-7 (2006)
2006
Show Abstract
The coordinated action of histone acetyltransferases (HATs) and ATP-dependent chromatin remodeling enzymes in promoter-dependent transcription initiation represents a paradigm for how epigenetic information regulates gene expression. However, little is known about how such enzymes function during transcription elongation. Here, we investigated the role of RSC, a bromodomain-containing ATPase, in nucleosome transcription in vitro. Purified S. cerevisiae RNA polymerase II (Pol II) arrests at two primary locations on a positioned mononucleosome. RSC stimulates passage of Pol II through these sites. The function of RSC in elongation requires the energy of ATP hydrolysis. Moreover, the SAGA and NuA4 HATs strongly stimulated RSC's effect on elongation. The stimulation correlates closely with acetyl-CoA-dependent recruitment of RSC to nucleosomes. Thus, RSC can recognize acetylated nucleosomes and facilitate passage of Pol II through them. These data support the view that histone modifications regulate accessibility of the coding region to Pol II. | | | 17081996
 |
Acetylated histone H4 is reduced in human gastric adenomas and carcinomas
Ono, S, et al
J Exp Clin Cancer Res, 21:377-82 (2002)
2002
| | | 12385581
 |