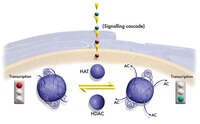
Coordinated regulation of active and repressive histone methylations by a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans.
Lin, H; Wang, Y; Wang, Y; Tian, F; Pu, P; Yu, Y; Mao, H; Yang, Y; Wang, P; Hu, L; Lin, Y; Liu, Y; Xu, Y; Chen, CD
Cell research
20
899-907
2010
Show Abstract
H3K9me2 and H3K27me2 are important epigenetic marks associated with transcription repression, while H3K4me3 is associated with transcription activation. It has been shown that active and repressive histone methylations distribute in a mutually exclusive manner, but the underlying mechanism was poorly understood. Here we identified ceKDM7A, a PHD (plant homeodomain)- and JmjC domain-containing protein, as a histone demethylase specific for H3K9me2 and H3K27me2. We further demonstrated that the PHD domain of ceKDM7A bound H3K4me3 and H3K4me3 co-localized with ceKDM7A at the genome-wide level. Disruption of the PHD domain binding to H3K4me3 reduced the demethylase activity in vivo, and loss of ceKDM7A reduced the expression of its associated target genes. These results indicate that ceKDM7A is recruited to the promoter to demethylate H3K9me2 and H3K27me2 and activate gene expression through the binding of the PHD domain to H3K4me3. Thus, our study identifies a dual-specificity histone demethylase and provides novel insights into the regulation of histone methylation. | 20567262
 |
The ciliary smooth muscle electrotransfer: basic principles and potential for sustained intraocular production of therapeutic proteins.
Elodie Touchard,Laura Kowalczuk,Carole Bloquel,Marie-Christine Naud,Pascal Bigey,Francine Behar-Cohen
The journal of gene medicine
12
2010
Show Abstract
We have developed a nonviral gene therapy method based on the electrotransfer of plasmid in the ciliary muscle. These easily accessible smooth muscle cells could be turned into a biofactory for any therapeutic proteins to be secreted in a sustained manner in the ocular media. | 21105151
 |
Expression of stress proteins and lymphocyte reactivity in heterotopic cardiac allografts undergoing cellular rejection.
J Qian,R Moliterno,M A Donovan-Peluso,K Liu,J Suzow,L Valdivia,F Pan,R J Duquesnoy
Transplant immunology
3
1995
Show Abstract
This report addresses the concept that, during rejection, the allograft undergoes a stress response which leads to an increased expression of stress proteins, also called heat shock proteins (hsp), and the recruitment and activation of hsp-reactive lymphocytes. Recent studies in our laboratory have provided evidence that hsp-reactive T-cells are present in cardiac allografts undergoing rejection. In this study, an MHC incompatible heterotopic heart allograft model (ACI into LEW) was chosen to analyse the kinetics of hsp expression during the development of rejection. Allografts and syngrafts (LEW into LEW) were harvested every day during the first 5 days post-transplant. Immunoblot analysis of proteins extracted from graft stromal tissues was done with murine monoclonal antibodies (mAb) against various mammalian hsp. Proliferation studies were done to determine hsp reactivity of graft-infiltrating lymphocytes on different days post-transplant. Three types of stressful stimuli appeared to increase hsp expression in the allograft. The first was a physiological stress secondary to the trauma of the transplant procedure and ischaemia/reperfusion injury and this would occur in allogeneic and syngeneic grafts. During the first day after transplantation, both types of grafts showed higher expression of hsp72 and grp78 and to a lesser extent, hsp60 and grp75. On the second and third day, the expression of grp78 and grp96 was higher in allografts than in syngrafts and this may reflect an immunologically mediated stress response in the allograft when infiltrating hsp-reactive lymphocytes became first detectable in the allograft. The third type of stress appeared related to the inflammatory process associated with rejection. On the fourth and fifth day post-transplant, the allografts showed strong expression of at least five proteins of lower molecular mass reacting with hsp-specific mAbs; namely, approximately 40 kDa (detected by anti-hsp60), approximately 30 kDa (by anti-hsp72), approximately 45 kDa and approximately 32 kDa (by anti-hsp72 + hsc73), and approximately 50 kDa (by anti-grp78). At that time, the allograft began to show progressive inflammatory changes and tissue damage. The appearance of lower molecular mass hsp-crossreactive proteins might reflect a degradation of hsps which had increased expression earlier during the post-transplant period. This process may generate large quantities of hsp-derived peptides which may be presented by MHC molecules to graft-infiltrating T-cells. Another interpretation of the strong expression of lower molecular bands in later allografts is that they represent other stress proteins that crossreact with antibodies against hsp60 and hsp70 family members.(ABSTRACT TRUNCATED AT 400 WORDS) | 7582902
 |