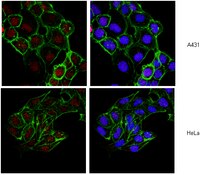
The hydroxylation activity of Jmjd6 is required for its homo-oligomerization.
Gang Han,Jiajia Li,Yiqin Wang,Xia Li,Hailei Mao,Yifan Liu,Charlie Degui Chen
Journal of cellular biochemistry
113
2012
Show Abstract
Jumonji C-terminal (JmjC) domain-containing proteins are protein hydroxylases and histone demethylases that control gene expression. Jumonji domain-containing protein 6 (Jmjd6) is indispensable for embryonic development and has both histone arginine demethylase and lysyl-hydroxylase activities. The protein undergoes post-translational homo-oligomerization, but the underlying mechanism remains unknown. In this study, we examined the enzymatic activity of Jmjd6 and uncovered the mechanism underlying its homo-oligomerization. An in vitro enzymatic assay monitored by matrix-assisted laser desorption-ionization time-of-flight (MALDI-TOF) mass spectrometry indicates that Jmjd6 is unable to remove the methyl group from histone arginine residues but can hydroxylate the histone H4 tail at lysine residues in a 2-oxoglutarate (2-OG)- and Fe (II)-dependent manner. A mutational analysis reveals that the homo-oligomerization of Jmjd6 requires its enzymatic activity and the N- and C-termini. Using an in vitro enzymatic assay, we further demonstrate that Jmjd6 can hydroxylate its N-terminus but not its C-terminus. In summary, we did not detect arginine demethylase activity for Jmjd6, but we did confirm that it could catalyze the lysyl-hydroxylation of histone peptides. In addition, we demonstrated that the homo-oligomerization of Jmjd6 requires its own enzymatic activity and the N- and C-termini. We propose that Jmjd6 forms intermolecular covalent bonds between its N- and C-termini via autohydroxylation. | 22189873
 |
Maternally transmitted partial direct tandem duplication of mitochondrial DNA associated with diabetes mellitus.
D R Dunbar,P A Moonie,R J Swingler,D Davidson,R Roberts,I J Holt
Human molecular genetics
2
1993
Show Abstract
Mitochondrial DNA from a 38 year old male with diabetes mellitus and features of mitochondrial dysfunction was analysed and shown to include a population with a partial duplication. The partially duplicated mitochondrial DNA molecules were evident in both muscle and blood. The region of mitochondrial DNA duplicated includes the origin of heavy strand replication, but not the light strand origin. This patient has features in common with other cases of partial direct tandem duplications and with a family which was reported to harbour a 10.4 kb mtDNA deletion. Initial restriction enzyme analysis of our case produced results consistent with a partial deletion of mitochondrial DNA. This leads us to propose that the rarity of reports of partial mitochondrial DNA duplications may stem in part from the classification of such mutants as partial deletions. | 8268914
 |