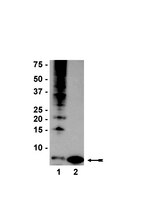
In vivo monoubiquitination of anaplerotic phosphoenolpyruvate carboxylase occurs at Lys624 in germinating sorghum seeds.
Ruiz-Ballesta, I; Feria, AB; Ni, H; She, YM; Plaxton, WC; Echevarría, C
Journal of experimental botany
65
443-51
2014
Show Abstract
Phosphoenolpyruvate carboxylase (PEPC; EC 4.1.1.31) is an important cytosolic regulatory enzyme that plays a pivotal role in numerous physiological processes in plants, including seed development and germination. Previous studies demonstrated the occurrence of immunoreactive PEPC polypeptides of ~110 kDa and 107 kDa (p110 and p107, respectively) on immunoblots of clarified extracts of germinating sorghum (Sorghum bicolor) seeds. In order to establish the biochemical basis for this observation, a 460 kDa PEPC heterotetramer composed of an equivalent ratio of p110 and p107 subunits was purified to near homogeneity from the germinated seeds. Mass spectrometry established that p110 and p107 are both encoded by the same plant-type PEPC gene (CP21), but that p107 was in vivo monoubiquitinated at Lys624 to form p110. This residue is absolutely conserved in vascular plant PEPCs and is proximal to a PEP-binding/catalytic domain. Anti-ubiquitin IgG immunodetected p110 but not p107, whereas incubation with a deubiquitinating enzyme (USP-2 core) efficiently converted p110 into p107, while relieving the enzyme's feedback inhibition by L-malate. Partial PEPC monoubiquitination was also detected during sorghum seed development. It is apparent that monoubiquitination at Lys624 is opposed to phosphorylation at Ser7 in terms of regulating the catalytic activity of sorghum seed PEPC. PEPC monoubiquitination is hypothesized to fine-tune anaplerotic carbon flux according to the cell's immediate physiological requirements for tricarboxylic acid cycle intermediates needed in support of biosynthesis and carbon-nitrogen interactions. | | 24288181
 |
The degradation of p53 and its major E3 ligase Mdm2 is differentially dependent on the proteasomal ubiquitin receptor S5a.
Sparks, A; Dayal, S; Das, J; Robertson, P; Menendez, S; Saville, MK
Oncogene
33
4685-96
2014
Show Abstract
p53 and its major E3 ligase Mdm2 are both ubiquitinated and targeted to the proteasome for degradation. Despite the importance of this in regulating the p53 pathway, little is known about the mechanisms of proteasomal recognition of ubiquitinated p53 and Mdm2. In this study, we show that knockdown of the proteasomal ubiquitin receptor S5a/PSMD4/Rpn10 inhibits p53 protein degradation and results in the accumulation of ubiquitinated p53. Overexpression of a dominant-negative deletion of S5a lacking its ubiquitin-interacting motifs (UIM)s, but which can be incorporated into the proteasome, also causes the stabilization of p53. Furthermore, small-interferring RNA (siRNA) rescue experiments confirm that the UIMs of S5a are required for the maintenance of low p53 levels. These observations indicate that S5a participates in the recognition of ubiquitinated p53 by the proteasome. In contrast, targeting S5a has no effect on the rate of degradation of Mdm2, indicating that proteasomal recognition of Mdm2 can be mediated by an S5a-independent pathway. S5a knockdown results in an increase in the transcriptional activity of p53. The selective stabilization of p53 and not Mdm2 provides a mechanism for p53 activation. Depletion of S5a causes a p53-dependent decrease in cell proliferation, demonstrating that p53 can have a dominant role in the response to targeting S5a. This study provides evidence for alternative pathways of proteasomal recognition of p53 and Mdm2. Differences in recognition by the proteasome could provide a means to modulate the relative stability of p53 and Mdm2 in response to cellular signals. In addition, they could be exploited for p53-activating therapies. This work shows that the degradation of proteins by the proteasome can be selectively dependent on S5a in human cells, and that this selectivity can extend to an E3 ubiquitin ligase and its substrate. | Western Blotting | 24121268
 |
Osmotic stress changes the expression and subcellular localization of the Batten disease protein CLN3.
Getty, A; Kovács, AD; Lengyel-Nelson, T; Cardillo, A; Hof, C; Chan, CH; Pearce, DA
PloS one
8
e66203
2013
Show Abstract
Juvenile CLN3 disease (formerly known as juvenile neuronal ceroid lipofuscinosis) is a fatal childhood neurodegenerative disorder caused by mutations in the CLN3 gene. CLN3 encodes a putative lysosomal transmembrane protein with unknown function. Previous cell culture studies using CLN3-overexpressing vectors and/or anti-CLN3 antibodies with questionable specificity have also localized CLN3 in cellular structures other than lysosomes. Osmoregulation of the mouse Cln3 mRNA level in kidney cells was recently reported. To clarify the subcellular localization of the CLN3 protein and to investigate if human CLN3 expression and localization is affected by osmotic changes we generated a stably transfected BHK (baby hamster kidney) cell line that expresses a moderate level of myc-tagged human CLN3 under the control of the human ubiquitin C promoter. Hyperosmolarity (800 mOsm), achieved by either NaCl/urea or sucrose, dramatically increased the mRNA and protein levels of CLN3 as determined by quantitative real-time PCR and Western blotting. Under isotonic conditions (300 mOsm), human CLN3 was found in a punctate vesicular pattern surrounding the nucleus with prominent Golgi and lysosomal localizations. CLN3-positive early endosomes, late endosomes and cholesterol/sphingolipid-enriched plasma membrane microdomain caveolae were also observed. Increasing the osmolarity of the culture medium to 800 mOsm extended CLN3 distribution away from the perinuclear region and enhanced the lysosomal localization of CLN3. Our results reveal that CLN3 has multiple subcellular localizations within the cell, which, together with its expression, prominently change following osmotic stress. These data suggest that CLN3 is involved in the response and adaptation to cellular stress. | | 23840424
 |
Tissue-specific expression and post-translational modifications of plant- and bacterial-type phosphoenolpyruvate carboxylase isozymes of the castor oil plant, Ricinus communis L.
O'Leary, B; Fedosejevs, ET; Hill, AT; Bettridge, J; Park, J; Rao, SK; Leach, CA; Plaxton, WC
Journal of experimental botany
62
5485-95
2011
Show Abstract
This study employs transcript profiling together with immunoblotting and co-immunopurification to assess the tissue-specific expression, protein:protein interactions, and post-translational modifications (PTMs) of plant- and bacterial-type phosphoenolpyruvate carboxylase (PEPC) isozymes (PTPC and BTPC, respectively) in the castor plant, Ricinus communis. Previous studies established that the Class-1 PEPC (PTPC homotetramer) of castor oil seeds (COS) is activated by phosphorylation at Ser-11 and inhibited by monoubiquitination at Lys-628 during endosperm development and germination, respectively. Elimination of photosynthate supply to developing COS by depodding caused the PTPC of the endosperm and cotyledon to be dephosphorylated, and then subsequently monoubiquitinated in vivo. PTPC monoubiquitination rather than phosphorylation is widespread throughout the castor plant and appears to be the predominant PTM of Class-1 PEPC that occurs in planta. The distinctive developmental patterns of PTPC phosphorylation versus monoubiquitination indicates that these two PTMs are mutually exclusive. By contrast, the BTPC: (i) is abundant in the inner integument, cotyledon, and endosperm of developing COS, but occurs at low levels in roots and cotyledons of germinated COS, (ii) shows a unique developmental pattern in leaves such that it is present in leaf buds and young expanding leaves, but undetectable in fully expanded leaves, and (iii) tightly interacts with co-expressed PTPC to form the novel and allosterically-desensitized Class-2 PEPC heteromeric complex. BTPC and thus Class-2 PEPC up-regulation appears to be a distinctive feature of rapidly growing and/or biosynthetically active tissues that require a large anaplerotic flux from phosphoenolpyruvate to replenish tricarboxylic acid cycle C-skeletons being withdrawn for anabolism. | | 21841182
 |
The Caenorhabditis elegans Elongator complex regulates neuronal alpha-tubulin acetylation.
Solinger, JA; Paolinelli, R; Klöss, H; Scorza, FB; Marchesi, S; Sauder, U; Mitsushima, D; Capuani, F; Stürzenbaum, SR; Cassata, G
PLoS genetics
6
e1000820
2010
Show Abstract
Although acetylated alpha-tubulin is known to be a marker of stable microtubules in neurons, precise factors that regulate alpha-tubulin acetylation are, to date, largely unknown. Therefore, a genetic screen was employed in the nematode Caenorhabditis elegans that identified the Elongator complex as a possible regulator of alpha-tubulin acetylation. Detailed characterization of mutant animals revealed that the acetyltransferase activity of the Elongator is indeed required for correct acetylation of microtubules and for neuronal development. Moreover, the velocity of vesicles on microtubules was affected by mutations in Elongator. Elongator mutants also displayed defects in neurotransmitter levels. Furthermore, acetylation of alpha-tubulin was shown to act as a novel signal for the fine-tuning of microtubules dynamics by modulating alpha-tubulin turnover, which in turn affected neuronal shape. Given that mutations in the acetyltransferase subunit of the Elongator (Elp3) and in a scaffold subunit (Elp1) have previously been linked to human neurodegenerative diseases, namely Amyotrophic Lateral Sclerosis and Familial Dysautonomia respectively highlights the importance of this work and offers new insights to understand their etiology. | | 20107598
 |
Posttranslational modification of ataxin-7 at lysine 257 prevents autophagy-mediated turnover of an N-terminal caspase-7 cleavage fragment.
Shona Mookerjee,Theodora Papanikolaou,Stephan J Guyenet,Vanitha Sampath,Amy Lin,Cathy Vitelli,Francesco DeGiacomo,Bryce L Sopher,Sylvia F Chen,Albert R La Spada,Lisa M Ellerby
The Journal of neuroscience : the official journal of the Society for Neuroscience
29
2009
Show Abstract
Polyglutamine (polyQ) expansion within the ataxin-7 protein, a member of the STAGA [SPT3-TAF(II)31-GCN5L acetylase] and TFTC (GCN5 and TRRAP) chromatin remodeling complexes, causes the neurodegenerative disease spinocerebellar ataxia type 7 (SCA7). Proteolytic processing of ataxin-7 by caspase-7 generates N-terminal toxic polyQ-containing fragments that accumulate with disease progression and play an important role in SCA7 pathogenesis. To elucidate the basis for the toxicity of these fragments, we evaluated which posttranslational modifications of the N-terminal fragment of ataxin-7 modulate turnover and toxicity. Here, we show that mutating lysine 257 (K257), an amino acid adjacent to the caspase-7 cleavage site of ataxin-7 regulates turnover of the truncation product in a repeat-dependent manner. Modification of ataxin-7 K257 by acetylation promotes accumulation of the fragment, while unmodified ataxin-7 is degraded. The degradation of the caspase-7 cleavage product is mediated by macroautophagy in cell culture and primary neuron models of SCA7. Consistent with this, the fragment colocalizes with autophagic vesicle markers, and enhanced fragment accumulation increases in these lysosomal structures. We suggest that the levels of fragment accumulation within the cell is a key event in SCA7 neurodegeneration, and enhancing clearance of polyQ-containing fragments may be an effective target to reduce neurotoxicity in SCA7. Full Text Article | | 19955365
 |
Regulatory monoubiquitination of phosphoenolpyruvate carboxylase in germinating castor oil seeds.
Uhrig, RG; She, YM; Leach, CA; Plaxton, WC
The Journal of biological chemistry
283
29650-7
2008
Show Abstract
Phosphoenolpyruvate carboxylase (PEPC) is a tightly regulated enzyme situated at the core of plant C-metabolism. Although its anaplerotic role and control by allosteric effectors, reversible phosphorylation, and oligomerization have been well documented in the endosperm of developing castor oil seeds (COS), relatively little is known about PEPC in germinating COS. The initial phase of COS germination was accompanied by elevated PEPC activity and accumulation of comparable amounts of pre-existing 107-kDa and inducible 110-kDa immunoreactive PEPC polypeptides (p107 and p110, respectively). A 440-kDa PEPC heterotetramer composed of an equivalent ratio of non-phosphorylated p110 and p107 subunits was purified from germinated COS. N-terminal microsequencing, mass spectrometry, and immunoblotting revealed that both subunits arose from the same gene (RcPpc3) that encodes the p107 subunit of a phosphorylated 410-kDa PEPC homotetramer in developing COS but that p110 is a monoubiquitinated form of p107. Tandem mass spectrometry sequencing of a diglycinated tryptic peptide identified Lys-628 as p110's monoubiquitination site. This residue is conserved in vascular plant PEPCs and is proximal to a PEP-binding/catalytic domain. Incubation with a human deubiquitinating enzyme (USP-2 core) converted the p110:p107 PEPC heterotetramer into a p107 homotetramer while significantly reducing the enzyme's K(m)(PEP) and sensitivity to allosteric activators (hexose-Ps, glycerol-3-P) and inhibitors (malate, aspartate). Monoubiquitination is a non-destructive and reversible post-translational modification involved in the control of diverse processes such as transcription, endocytosis, and signal transduction. The current study demonstrates that tissue-specific monoubiquitination of a metabolic enzyme can also occur and that this modification influences its kinetic and regulatory properties. | Western Blotting | 18728004
 |
The ubiquitin-proteasome pathway and its role in cancer.
Mani, Aparna and Gelmann, Edward P
J. Clin. Oncol., 23: 4776-89 (2005)
2005
Show Abstract
Critical cellular processes are regulated, in part, by maintaining the appropriate intracellular levels of proteins. Whereas de novo protein synthesis is a comparatively slow process, proteins are rapidly degraded at a rate compatible with the control of cell cycle transitions and cell death induction. A major pathway for protein degradation is initiated by the addition of multiple 76-amino acid ubiquitin monomers via a three-step process of ubiquitin activation and substrate recognition. Polyubiquitination targets proteins for recognition and processing by the 26S proteasome, a cylindrical organelle that recognizes ubiquitinated proteins, degrades the proteins, and recycles ubiquitin. The critical roles played by ubiquitin-mediated protein turnover in cell cycle regulation makes this process a target for oncogenic mutations. Oncogenes of several common malignancies, for example colon and renal cell cancer, code for ubiquitin ligase components. Cervical oncogenesis by human papillomavirus is also mediated by alteration of ubiquitin ligase pathways. Protein degradation pathways are also targets for cancer therapy, as shown by the successful introduction of bortezomib, an inhibitor of the 26S proteasome. Further work in this area holds great promise toward our understanding and treatment of a wide range of cancers. | | 16034054
 |
The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle.
Hershko, A
Cell Death Differ., 12: 1191-7 (2005)
2005
Show Abstract
Owing to the intensive research activity on protein synthesis, little attention was paid in the 1950s and 1960s to protein degradation. However, work by my group and others between 1970 and 1990 led to the identification of the ubiquitin-dependent degradation system. We found that this system contains three types of enzymes: E1 ubiquitin--activating enzyme, E2 ubiquitin--carrier enzyme and E3 ubiquitin--protein ligase. The sequential action of these enzymes leads to conjugation of ubiquitin to proteins and then in most cases to their degradation. This review briefly tells the story of how this pathway was discovered describing the main findings that during the years allowed us to draw the complex picture we have now. | | 16094395
 |
Regulating the regulators: control of protein ubiquitination and ubiquitin-like modifications by extracellular stimuli.
Gao, Min and Karin, Michael
Mol. Cell, 19: 581-93 (2005)
2005
Show Abstract
Like many other posttranscriptional modifications, ubiquitination and conjugation of ubiquitin-like polypeptides to target proteins are tightly regulated by extracellular stimuli. In many cases, this regulation is dependent upon protein phosphorylation. The regulatory step affected by phosphorylation could involve either recognition of the substrate by an E3 ubiquitin ligase or the actual conjugation reaction. Regulation occurs through phosphorylation of either the substrates or the E3 ligases themselves. This review focuses on recent advances in understanding how extracellular stimuli modulate the attachment of ubiquitin and ubiquitin-like peptides to target proteins. | | 16137616
 |